guest post by David Tweed
The members of the Azimuth Project have been working on both predicting and understanding the El Niño phenomenon, along with writing expository articles. So far we’ve mostly talked about the physics and data of the El Niño, along with looking at one method of actually trying to predict El Niño events. Since there’s going to more data exploration using methods more typical of machine learning, it’s a good time to briefly describe the mindset and highlight some of differences between different kinds of predictive models. Here we’ll concentrate on the concepts rather than the fine details and particular techniques.
We also stress there’s not a fundamental distinction between machine learning (ML) and statistical modelling and inference. There are certainly differences in culture, background and terminology, but in terms of the actual algorithms and mathematics used there’s a great commonality. Throughout the rest of the article we’ll talk about ‘machine learning models’, but could equally have used ‘statistical models’.
For our purposes here, a model is any object which provides a systematic procedure for taking some input data and producing a prediction of some output. There’s a spectrum of models, ranging from physically based models at one end to purely data driven models at the other. As a very simple example, suppose you commute by car from your place of work to your home and you want to leave work in order to arrive homeat 6:30 pm. You can tackle this by building a model which takes as input the day of the week and gives you back a time to leave.
• There’s the data driven approach, where you try various leaving times on various days and record whether or not you get home by 6:30 pm. You might find that the traffic is lighter on weekend days so you can leave at 6:10 pm while on weekdays you have to leave at 5:45 pm, except on Wednesdays when you have to leave at 5:30pm. Since you’ve just crunched on the data you have no idea why this works, but it’s a very reliable rule when you use it to predict when you need to leave.
• There’s the physical model approach, where you attempt to infer how many people are doing what on any given day and then figure out what that implies for the traffic levels and thus what time you need to leave. In this case you find out that there’s a mid-week sports game on Wednesday evenings which leads to even higher traffic. This not only predicts that you’ve got to leave at 5:30 pm on Wednesdays but also lets you understand why. (Of course this is just an illustrative example; in climate modelling a physical model would be based upon actual physical laws such as conservation of energy, conservation of momentum, Boyle’s law, etc.)
There are trade-offs between the two types of approach. Data driven modelling is a relatively simple process. In contrast, by proceeding from first principles you’ve got a more detailed framework which is equally predictive but at the cost of having to investigate a lot of complicated underlying effects. Physical models have one interesting advantage: nothing in the data driven model prevents it violating physical laws (e.g., not conserving energy, etc) whereas a physically based model obeys the physical laws by design. This is seldom a problem in practice, but worth keeping in mind.
The situation with data driven techniques is analogous to one
of those American medication adverts: there’s the big message about how “using a data driven technique can change your life for the better” while the voiceover gabbles out all sorts of small print. The remainder of this post will describe some of the basic principles in that small print.
Preprocessing and feature extraction
There’s a popular misconception that machine learning works well when you simply collect some data and throw it into a machine learning algorithm. In practice that kind of approach often yields a model that is quite poor. Almost all successful machine learning applications are preceded by some form of data preprocessing. Sometimes this is simply rescaling so that different variables have similar magnitudes, are zero centred, etc.
However, there are often steps that are more involved. For example, many machine learning techniques have what are called ‘kernel variants’ which involve (in a way whose details don’t matter here) using a nonlinear mapping from the original data to a new space which is more amenable to the core algorithm. There are various kernels with the right mathematical properties, and frequently the choice of a good kernel is made either by experimentation or knowledge of the physical principles. Here’s an example (from Wikipedia’s entry on the support vector machine) of how a good choice of kernel can convert a not linearly separable dataset into a linearly separable one:
An extreme example of preprocessing is explicitly extracting features from the data. In ML jargon, a feature “boils down” some observed data into a value directly useful. For example, in the work by Ludescher et al that we’ve been looking at, they don’t feed all the daily time series values into their classifier but take the correlation between different points over a year as the basic features to consider. Since the individual days’ temperatures are incredibly noisy and there are so many of them, extracting features from them gives more useful input data. While these extraction functions could theoretically be learned by the ML algorithm, this is quite a complicated function to learn. By explicitly choosing to represent the data using this feature the amount the algorithm has to discover is reduced and hence the likelihood of it finding an excellent model is dramatically increased.
Limited amounts of data for model development
Some of the problems that we describe below would vanish if we had unlimited amounts of data to use for model development. However, in real cases we often have a strictly limited amount of data we can obtain. Consequently we need methodologies to address the issues that arise when data is limited.
Training sets and test sets
The most common way to work with collected data is to split it into a training set and a test set. The training set is used in the process of determining the best model parameters, while the test set—which is not used in any way in determining the those model parameters—is then used to see how effective the model is likely to be on new, unseen data. (The test and training sets need not be equally sized. There are some fitting techniques which need to further subdivide the training set, so that having more training than test data works out best.) This division of data acts to further reduce the effective amount of data used in determining the model parameters.
After we’ve made this split we have to be careful how much of the test data we scrutinise in any detail, since once it has been investigated it can’t meaningfully be used for testing again, although it can still be used for future training. (Examining the test data is often informally known as burning data.) That only applies to detailed inspection however; one common way to develop a model is to look at some training data and then train the model (also known as fitting the model) on that training data. It can then be evaluated on the test data to see how well it does. It’s also then okay to purely mechanically train the model on the test data and evaluate it on the training data to see how “stable” the performance is. (If you get dramatically different scores then your model is probably flaky!) However, once we start to look at precisely why the model failed on the test data—in order to change the form of the model—the test data has now become training data and can’t be used as test data for future variants of that model. (Remember, the real goal is to accurately predict the outputs for new, unseen inputs!)
Random patterns in small sample sets
Suppose we’re modelling a system which has a true probability distribution . We can’t directly observe this, but we have some samples
obtained from observation of the system and hence come from
. Clearly there are problems if we generate this sample in a way that will bias the area of the distribution we sample from: it wouldn’t be a good idea to get training data featuring heights in the American population by only handing out surveys in the locker rooms of basketball facilities. But if we take care to avoid sampling bias as much as possible, then we can make various kinds of estimates of the distribution that we think
comes from.
Let’s consider the estimate implied for
by some particular technique. It would be nice if
, wouldn’t it? And indeed many good estimators have the property that as the size of
tends to infinity
will tend to
. However, for finite sizes of
, and especially for small sizes,
may have some spurious detail that’s not present in
.
As a simple illustration of this, my computer has a pseudorandom number generator which generates essentially uniformly distributed random numbers between 0 and 32767. I just asked for 8 numbers and got
2928, 6552, 23979, 1672, 23440, 28451, 3937, 18910.
Note that we’ve got one subset of 4 values (2928, 6552, 1672, 3937) within the interval of length 5012 between 1540 and 6552 and another subset of 3 values (23440, 23979 and 28451) in the interval of length 5012 between 23440 and 28451. For this uniform distribution the expectation of the number of values falling within a given range of that size is about 1.2. Readers will be familiar with how the expectation of a random quantity for a small sample will have a large amount of variation around its value that only reduces as the sample size increases, so this isn’t a surprise. However, it does highlight that even completely unbiased sampling from the true distribution will typically give rise to extra ‘structure’ within the distribution implied by the samples.
For example, here are the results from one way of estimating the probability from the samples:
The green line is the true density while the red curve shows the probability density obtained from the samples, with clearly spurious extra structure.
Generalization
Almost all modelling techniques, while not necessarily estimating an explicit probability distribution from the training samples, can be seen as building functions that are related to that probability distribution.
For example, a ‘thresholding classifier’ for dividing input into two output classes will place the threshold at the optimal point for the distribution implied by the samples. As a consequence, one important aim in building machine learning models is to estimate the features that are present in the true probability distribution while not learning such fine details that they are likely to be spurious features due to the small sample size. If you think about this, it’s a bit counter-intuitive: you deliberately don’t want to perfectly reflect every single pattern in the training data. Indeed, specialising a model too closely to the training data is given the name over-fitting.
This brings us to generalization. Strictly speaking generalization is the ability of a model to work well upon unseen instances of the problem (which may be difficult for a variety of reasons). In practice however one tries hard to get representative training data so that the main issue in generalization is in preventing overfitting, and the main way to do that is—as discussed above—to split the data into a set for training and a set only used for testing.
One factor that’s often related to generalization is regularization, which is the general term for adding constraints to the model to prevent it being too flexible. One particularly useful kind of regularization is sparsity. Sparsity refers to the degree to which a model has empty elements, typically represented as 0 coefficients. It’s often possible to incorporate a prior into the modelling procedure which will encourage the model to be sparse. (Recall that in Bayesian inference the prior represents our initial ideas of how likely various different parameter values are.) There are some cases where we have various detailed priors about sparsity for problem specific reasons. However the more common case is having a ‘general modelling’ belief, based upon experience in doing modelling, that sparser models have a better generalization performance.
As an example of using sparsity promoting priors, we can look at linear regression. For standard regression with examples of
against
dimensional vectors
we’re considering the total error
while with the prior we’ve got
where are the coefficients to be fitted and
is the prior weight. We can see how the prior weight affects the sparsity of the
s:
On the -axis is
while the
-axis is the coefficient value. Each line represents the value of one particular coefficient as
increases. You can see that for very small
– corresponding to a very weak prior – all the weights are non-zero, but as it increases – corresponding to the prior becoming stronger – more and more of them have a value of 0.
There are a couple of other reasons for wanting sparse models. The obvious one is speed of model evaluation, although this is much less significant with modern computing power. A less obvious reason is that one can often only effectively utilise a sparse model, e.g., if you’re attempting to see how the input factors should be physically modified in order to affect the real system in a particular way. In this case we might want a good sparse model rather than an excellent dense model.
Utility functions and decision theory
While there are some situations where a model is sought purely to develop knowledge of the universe, in many cases we are interested in models in order to direct actions. For example, having forewarning of El Niño events would enable all sorts of mitigation actions. However, these actions are costly so they shouldn’t be undertaken when there isn’t an upcoming El Niño. When presented with an unseen input the model can either match the actual output (i.e., be right) or differ from the actual output (i.e., be wrong). While it’s impossible to know in advance if a single output will be right or wrong – if we could tell that we’d be better off using that in our model – from the training data it’s generally possible to estimate the fractions of predictions that will be right and will be wrong in a large number of uses. So we want to link these probabilities with the effects of actions taken in response to model predictions.
We can do this using a utility function and a loss
function. The utility maps each possible output to a numerical value proportional to the benefit from taking actions when that output was correctly anticipated. The loss maps outputs to a number proportional to the loss from the actions when the output was incorrectly predicted by the model. (There is evidence that human beings often have inconsistent utility and loss functions, but that’s a story for another day…)
There are three common ways the utility and loss functions are used:
• Maximising the expected value of the utility (for the fraction where the prediction is correct) minus the
expected value of the loss (for the fraction where the prediction is incorrect).
• Minimising the expected loss while ensuring that the expected utility is at least some
value
• Maximising the expected utility while ensuring that the expected loss is at most some
value.
Once we’ve chosen which one we want, it’s often possible to actually tune the fitting of the model to optimize with respect to that criterion.
Of course sometimes when building a model we don’t know enough details of how it will be used to get accurate utility and loss functions (or indeed know how it will be used at all).
Inferring a physical model from a machine learning model
It is certainly possible to take a predictive model obtained by machine learning and use it to figure out a physically based model; this is one way of performing data mining. However in practice there are a couple of reasons why it’s necessary to take some care when doing this:
• The variables in the training set may be related by some
non-observed latent variables which may be difficult to reconstruct without knowledge of the physical laws that are in play. (There are machine learning techniques which attempt to reconstruct unknown latent variables but this is a much more difficult problem than estimating known but unobserved latent variables.)
• Machine learning models have a maddening ability to find variables that are predictive due to the way the data was gathered. For example, in a vision system aimed at finding tanks all the images of tanks were taken during one day on a military base when there was accidentally a speck of grime on the camera lens, while all the images of things that weren’t tanks were taken on other days. A neural net cunningly learned that to decide if it was being shown a tank it should look for the shadow from the grime.
• It’s common to have some groups of very highly correlated input variables. In that case a model will generally learn a function which utilises an arbitrary linear combination of the correlated variables and an equally good model would result from using any other linear combination. (This is an example of the statistical problem of ‘identifiability’.) Certain sparsity encouraging priors have the useful property of encouraging the model to select only one representative from a group of correlated variables. However, even in that case it’s still important not to assign too much significance to the particular division of model parameters in groups of correlated variables.
• One can often come up with good machine learning models even when physically important variables haven’t been collected in the training data. A related issue is that if all the training data is collected from a particular subspace factors that aren’t important there won’t be found. For example, if in a collision system to be modelled all data is collected about low speeds the machine learning model won’t learn about relativistic effects that only have a big effect at a substantial fraction of the speed of light.
Conclusions
All of the ideas discussed above are really just ways of making sure that work developing statistical/machine learning models for a real problem is producing meaningful results. As Bob Dylan (almost) sang, “to live outside the physical law, you must be honest; I know you always say that you agree”.




thanks for the post!
I don’t know much about machine learning techniques, so my question may sound strange. You wrote that:
However Wikipedia writes:
This sounds to me as if the goal for support vector machines is to always construct hyper planes and that this can be achieved even if the kernel is nonlinear. Does the “kernel trick”-transform (construction of the feature map) involve the choice of parameters? Do you mean that counting measure? The Wikipedia explanation is rather too cryptic for me for getting a quick insight.
Since the article is such a whirlwind tour intended just to briefly mention some concepts that may come up in more detail it later blogs, it’s not surprising it’s hard to follow! :S
Here’s a concrete, toy example that may clarify things. Suppose we’ve got 1 point from class A at and 4 points from class B at
and 4 points from class B at  . If you draw this it’s easy to see there’s no straight line where all the B points are on the other side to the A point. If we map each point to
. If you draw this it’s easy to see there’s no straight line where all the B points are on the other side to the A point. If we map each point to  then we end up with just two distinct points and we find that
then we end up with just two distinct points and we find that  is the “maximum margin” straight line with A on one side and the B’s on the other in the new space. (We were freakily lucky that it’s an incredibly simple map and it doesn’t increase the number of dimensions, neither of which is true in general.)
is the “maximum margin” straight line with A on one side and the B’s on the other in the new space. (We were freakily lucky that it’s an incredibly simple map and it doesn’t increase the number of dimensions, neither of which is true in general.)
All we theoretically need is the straight line in the new space, but we can substitute in to see that the points on the line correspond to points where in the original space: the straight line boundary corresponds to a “nonlinear” circular one in the original space! Note that while figuring out the line parameters in the new space is mechanical, choosing
in the original space: the straight line boundary corresponds to a “nonlinear” circular one in the original space! Note that while figuring out the line parameters in the new space is mechanical, choosing  happened outside the algorithm, which is why (stretching the point a little!) I gave it as an example of preprocessing. (Choosing, say,
happened outside the algorithm, which is why (stretching the point a little!) I gave it as an example of preprocessing. (Choosing, say,  won’t make these points linearly separable.)
won’t make these points linearly separable.)
(The clever bits of support vector machines are doing all this very efficiently by not avoiding explicitly mentioning nonlinear mapping.)
thanks for the explanation.
Unfortunately I am not sure if I have fully understood what you mean with preprocessing. Do you mean that the algorithm goes through various cases as seems to be indicated by Wikipedia, but then where is the choice ? or is this preprocessing supposed to be done by a human?
By that whats written in Wikipedia I can’t really understand how exactly that “kernel trick” works and Wikipedia wrote:
Does the “may be” extend to the “transformed space high dimensional” and thus in particular could also include lower dimensional? What would be the kernel in your case? (wikipedia writes only cryptically that
where it is not even clear here to me what they mean with “dual” in this case.
Firstly, it’s perhaps relevant to clarify that kernel functions are an “optimization” of the computation process and that the fundamental picture can be seen just in terms of the nonlinear map . The kernel and nonlinear map are always related by
. The kernel and nonlinear map are always related by
Knowing you can find
you can find 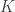 directly, while tells you how to go from a suitable
directly, while tells you how to go from a suitable 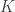 to
to  . For the example in my comment
. For the example in my comment
with corresponding kernel
But there’s nothing special about that : it could be
: it could be
or any other non-linear map. Different mappings will do better or worse in making the data separable using a hyperplane.
To see about the issue of choice, let’s look at the series of steps (in theory):
1. You give me a data set and ask for an SVM model.
2. I decide on a mapping (maybe after looking at the data).
(maybe after looking at the data).
3. I can stick the mapping and the data set into a standard SVM software package. In theory it applies to “preprocess” the dataset and then fits a maximum margin hyperplane to that transformed dataset.
to “preprocess” the dataset and then fits a maximum margin hyperplane to that transformed dataset.
4. I look at the performance of the classifier. If it’s not very good I might return to step 2 and try a different mapping.
5. I give you back the parameters for this SVM classifier.
The confusing bit comes because in 3 the package doesn’t literally apply to the dataset, instead it uses the kernel function to essentially do the
to the dataset, instead it uses the kernel function to essentially do the  mapping and the dot product using the one function
mapping and the dot product using the one function 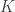 . It’s also the case that frequently you don’t choose
. It’s also the case that frequently you don’t choose  but go straight to choosing
but go straight to choosing 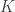 .
.
thanks for the reply.
If I understand correctly Wikipedia writes that in case that the data is linearly separable the kernel would be (after that ominous “dualization”): . So in the example that would be
. So in the example that would be  , if the set of points would be linearly separable, which it isn’t, that’s why you choose
, if the set of points would be linearly separable, which it isn’t, that’s why you choose  . Is that right? However if I am able to write a kernel as a dot-product
. Is that right? However if I am able to write a kernel as a dot-product  that doesn’t seem to guarantee that the corresponding set is automatically linearly separable. You wrote in that direction
that doesn’t seem to guarantee that the corresponding set is automatically linearly separable. You wrote in that direction
And so in the moment I say Yes. But….like if I choose to be the identity I will have the same problems as before. That is Mercer’s theorem doesn’t seem to be enough. There must be something, which tells you when that points get linearly separable (for the corresponding
to be the identity I will have the same problems as before. That is Mercer’s theorem doesn’t seem to be enough. There must be something, which tells you when that points get linearly separable (for the corresponding  ). What you seem to propose is that this is done by trial and error. And this “trial and error” prodedure is what you call the choice in preprocessing? Did I understand you right?
). What you seem to propose is that this is done by trial and error. And this “trial and error” prodedure is what you call the choice in preprocessing? Did I understand you right?
You’ve pretty much got it. The duality that wikipedia mentions is “just” one of the constrained optimization dualities, so while it’s still a strange fact it’s not something unique to the SVM.
thanks for the reply.
Somehow I feel still somewhat uneasy about that dualization methods and that trial and error methods. How is the trial and error method computationally implemented? Is that fast enough?
Wikipedia writes on the page of neural nets that:
and in fact there are even challenges on that topic:
Since as I understood the whole inner city of London is scanned with CCTV cameras and individuals are traced by computers for behaving against the norm – like rakishly put: deviate from their way between two subway stations for e.g. peeing against a lantern – the trial and error algorithms must be quite fast, at least in computer vision.
I’ve been a bit snowed under so haven’t got time to write a longer reply, but you might find this link shows some of the practical issues in using a kernel method (in this case Kernel PCA).
[…] Science, Models and Machine Learning | Azimuth. […]
I am glad that you focus heavily on cross-validation in your posts. I am often surprised by how often cross-validation is not used, although I would also argue that there are settings when it shouldn’t be used. You’ve side-stepped this issue a bit by defining models as predictive beasts, but there are definitely cases when you just want to focus on descriptive modeling without paying too much attention to prediction.
In Kuhn’s terminology, this would be the prescientific or pre-paradigm stage (in the machine learning setting, it might be the preprocessing and feature extraction stage). Here you are searching for a language with which you want to describe your goals (i.e. what to predict accurately) and the basic theoretical quantities of interest. If you become too prediction focused at this stage, it is easy to chase yourself into a corner. Imagine if physicists were concerned with predicting the shape or the color or the saltiness of an individual electron. From a folk theory perspective it seems like potentially reasonable question to ask. But if predicting saltiness was the measure of progress and we stuck dogmatically to it, we could easily have given up or been left with nonsense theories.
To bring this back to environmentalism: these seems to be at least some people that believe that environmentalism is still in its pre-paradigm stage. I am not informed enough to judge their assessment, but in such settings it seems that side-stepping the prediction-vs-description discussion might not be productive.
In general I enjoyed your post and my complaints are stemming not from what you wrote but what you had to omit: i.e. more connections to the philosophy of science.
I’m glad you liked the article. I read your blog and I think we have pretty complementary views.
I think that, whether you’re doing predictive or descriptive modelling, you want to try to keep your model relating to the original system rather than the particular set of data you got out of the system. Cross validation is one way of doing that, but so is having so much data that there’s no likelihood of spurious features, applying your prior views (I’m a fully signed up Bayesian!), putting a strong sparsity constraint on the model, etc.
With regard to cross validation in particular, there are certainly cases where it may not buy you anything, so there’s no point in doing it. For me the really important point is that if you claim to have done cross validation you do need not to have intensely scrutinised your test set, otherwise you’re not putting up independent evidence.
This is a good point, and I will have to think more carefully about it. The only response that springs to mind quickly is if the ‘original system’ is not a well defined term (for instance, the realist stance cannot be easily taken, as is the case sometimes in history) or if your method of getting data or ways of describing the data you got is an important focus. Both of these concerns seem to be important in the pre-paradigm stage.
I can’t say that I am, although I am definitely not a frequentist. My real issue is with how to properly handle non-determinism.
Can you expand on this a bit? Especially in the context of the quote-of-yours I opened with and also in the case of predictive modeling.
When I say “may not buy you anything”, all I mean is that cross validation isn’t doing very much if your data set is so small (relative to the complexity of the problem) that you’d be doing it with tiny training and validation sets, or as you mention when you have no idea which variables ought to considered outputs of the others (or even make sense. In the quote I’m just weakly arguing against the idea that there’s some “mechanical checklist” that you should follow when applying machine learning; people should be considering what they need to do to in their particular situation with their data.
There’s a parallel discussion going on about this post over on G+, and Neil Lawrence wrote:
Deen Abiola wrote:
Thomas Dietterich wrote:
The link to https://www.ceremade.dauphine.fr/~peyre/numerical-tour/tours/optim_8_homotopy/index_04.png is broken